Introduction
In bioinformatics, the availability of open-source software and resources has been a game-changer. Researchers can now access and analyze vast amounts of biological data using powerful, freely available computational tools. This has democratized the field and allowed researchers from all over the world to collaborate and contribute to the advancement of biological knowledge.
This document will discuss the benefits of leveraging open-source bioinformatics resources and provide examples of how researchers can use them to enhance their research.
Leveraging open-source bioinformatics databases can be incredibly valuable for various research and analysis purposes in biology and genetics. These databases provide access to vast amounts of biological data, allowing researchers to explore and interpret information related to genes, proteins, genomes, pathways, and more.
Some of the key databases are NCBI, Ensembl, TCGA, GEO (Gene expression Omnibus), which are used widely by researchers and scientists worldwide and are being updated with increased frequency. These databases, among many others, serve as valuable resources for scientists and bioinformaticians. Leveraging these open-source bioinformatics databases can aid in data analysis, hypothesis generation, and gaining insights into biological processes, ultimately advancing research and discovery in bioinformatics.
Open-source bioinformatics repositories provide a wealth of information that can be used in biological research. Here are some strategies for effectively using these resources:
1. Identify the right resource: With the plethora of open-source bioinformatics resources available, it is important to identify the right tool /data that suits your research needs. It is important to understand the nature of your research problem and the type of data you are working with to identify the right resource. For example, if you are working with next-generation sequencing data, tools like Galaxy, NCBI BLAST, and Bioconductor can be useful.
2. Familiarize yourself with the resource: Once you have identified the right resource, familiarize yourself with it. Learn about its features, functionalities, and limitations. Take advantage of online tutorials and documentation to better understand the tool/data.
3. Collaborate with others: Collaborating with other researchers using similar resources can be helpful and can be achieved through online forums, mailing lists, and social media. Collaborating with others can provide valuable insights and solutions to problems that you may encounter.
4. Keep up-to-date: Bioinformatics resources are constantly evolving. Keeping up-to-date with the latest developments and updates is important to ensure you use the most current and accurate information.
5. Participate in the community: Participating in the open-source Bioinformatics community can provide valuable feedback and insights.
Description of ASD
Autism Spectrum Disorder [Mendelian inheritance in man [(MIM) 209850], commonly referred to as ASD is a cluster of neurodevelopmental and neuropsychiatric abnormalities which manifest in individuals with deficiencies in social interactions, communications, repetitive behaviour and developmental issues which arises early (first two years) in the life of an individual. ASD is categorized as syndromic if the affected individual is associated with another neurological disorder and non-syndromic if ASD occurs without any other neurological disorder.
ASD is now prevalent in many countries and is present in 1% of the children population. Individuals with ASD are afflicted with cognitive function impairments, language development and intellectual disability (ID). The term spectrum points to a cluster of wide-ranging symptoms and disorders exhibited by individuals with ASD. The symptoms can be clinical and behavioural, which are ingused for diagnostic purposes.
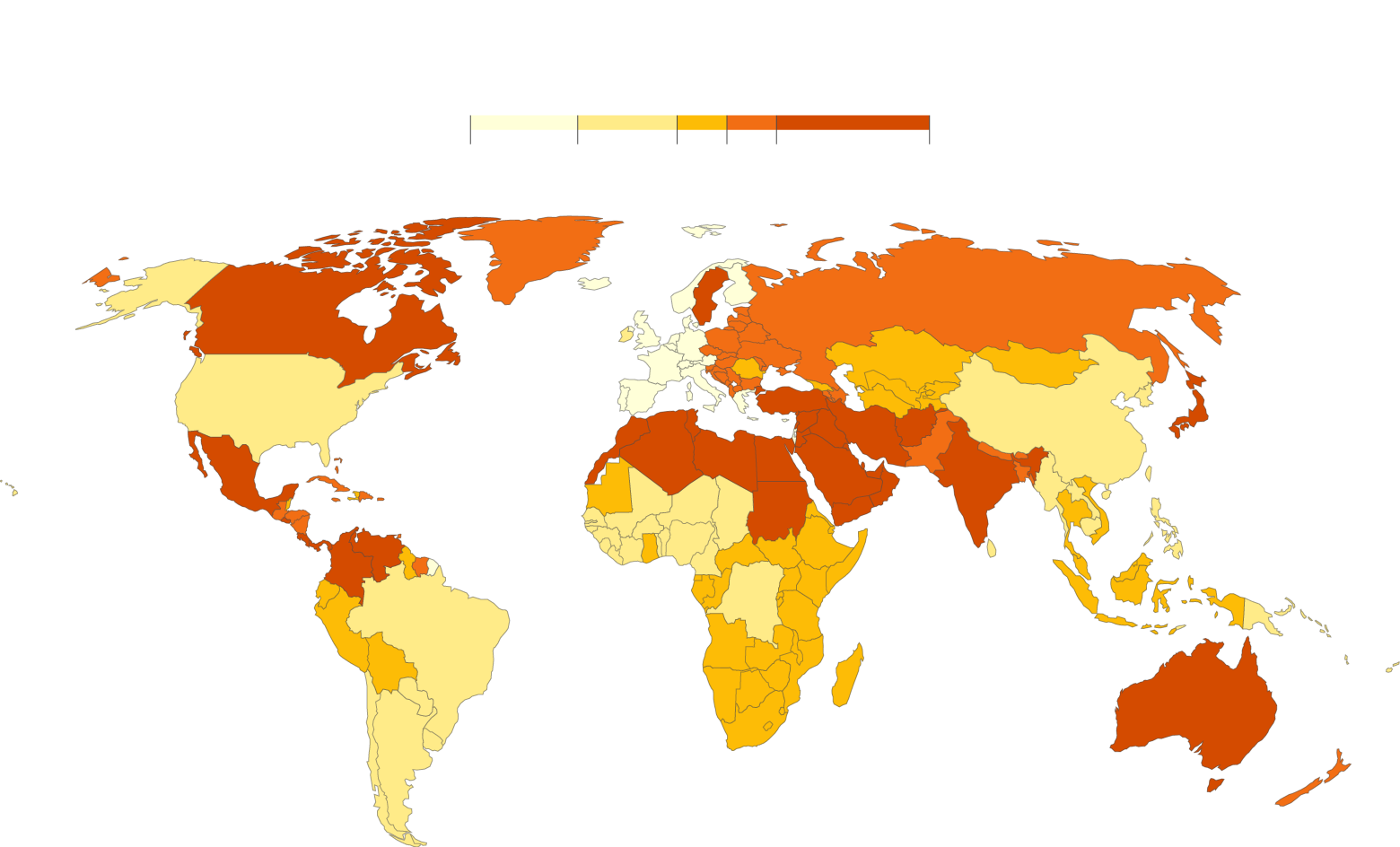
Figure 1. ASD prevalence around the world. [source: https://www.axios.com/2018/09/27/most-of-the-worlds-autistic-children-live-in-countries-with-few-resources]
The figure above (Figure 1.) shows the prevalence of ASD around the world. The figure is one of the best examples of merging public data sources with genetics data to show the distribution of the disease in various countries.
Open-Source Bioinformatics & ASD resources
The research done on ASD for the past 10 years with the advent of the ‘Omics’ era has generated a huge volume of data both metadata and bioinformatics data which includes, potential list of genes, gene sequences, ASD data repositories, bioinformatics analysis tools which have the power and versatility to handle large volume of data. Another simple data analysis shows the trend of discovery in ASD. The analysis was done using publicly available research which are housed in public data repositories such NCBI, EBI and other publicly funded databases.
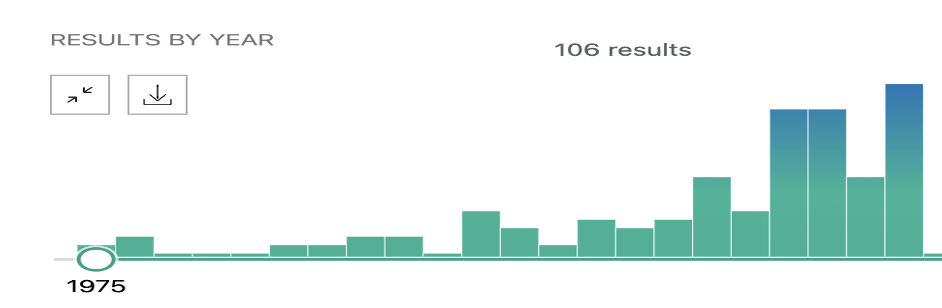
Figure 2. Literature analysis from the year 1975 till 1996. [source : NCBI – Keyword used: ASD]
The figure (Figure 2.) shows that only a handful of researchers were working on ASD simply based on the publication history from 1970 – 1996 for which data was sourced from NCBI and also other public data sources. In contrast, from 1996 till the year 2022, the below figure (Figure 3.) shows the explosion of research which was generated due to the availability of Bioinformatics & “Omics” data. Almost ~1500 research papers were published within this time frame which again is due to the omics data generated due to the advancement in sequencing technologies and public funding to make the data available to the public and research.
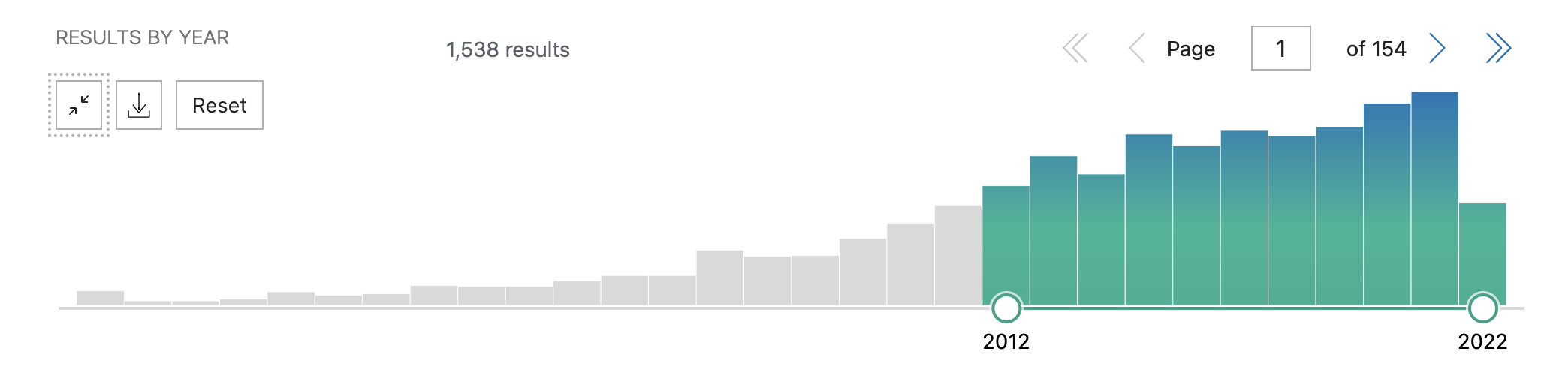
Figure 3. Literature analysis from the year 1996 till 2022. [source : NCBI - add parameters ]
The public & open-source data repositories available for ASD research are given below, with URL’s provided for easy reference and access. The 1st table (Table 1) has links to Bioinformatics data repositories which also contain information regarding ASD. The 2nd table is specific to ASD-related data sources, which are helpful to ASD researchers (Table 2).
Table 1: Open-Source Bioinformatics data repositories and databases
| S.No | Name | URL |
|---|---|---|
| 1 | NCBI – National Center for Bioinformatics information | https://www.ncbi.nlm.nih.gov |
| 2 | EBI – European Bioinformatics Institute | https://www.ebi.ac.uk |
| 3 | GEO – Gene Expression Omnibus | https://www.ncbi.nlm.nih.gov/geo/ |
| 4 | DECIPHER - | https://www.deciphergenomics.org |
| 5 | GTex – Genotype -Tissue Expression Portal | https://gtexportal.org/home/ |
| 6 | OpenTargets | https://www.opentargets.org |
| 7 | OmicsDI | https://www.omicsdi.org |
Table 2: Open-source Autism specific data repositories and databases
| S.No | Name | URL |
|---|---|---|
| 1 | SFARI – Simons Foundation Autism Research Initiative | https://www.sfari.org |
| 2 | AutDB – Autism Database | http://www.mindspec.org/autdb.html |
| 3 | NDAR – National Database of Autism Research | https://nda.nih.gov |
| 4 | Autism Chromosomal Rearrangement Database | http://projects.tcag.ca/autism/ |
| 5 | CNV database for Autism Research | https://www.med.kobe-u.ac.jp/asddb/ |
| 6 | Varicarta | https://varicarta.msl.ubc.ca/index |
The generation of genome level data for ASD patients and families has added another dimension in the data realm. Publicly available WGS samples (individual/family) along with tissue and organ specific Omics data has made ASD research more insightful and has provided more in-depth results in the field of ASD.
The most common source is the various databases of NCBI which house the largest collection of public sequence data for analysis. There is a search capability and advanced parameters which help in filtering the required data.In addition, other public datasets such as gnomAD at the Broad Institute are accessible to researchers for analysis. gnomAD data is available for download through Google Cloud Public Datasets, the Registry of Open Data on AWS, and Azure Open Datasets.
The NDAR database listed above contains multidisciplinary biomolecular data, including exome sequencing, brain imaging, and clinical diagnostic data. The level of heterogeneity in ASD research is unprecedented and can provide a better understanding of this complex disease. The Aut10K is a genome mapping study database which uses Google Cloud to manage, analyze, and disseminate its data, which provides new opportunities for broader public access.
The availability of WGS data has provided an additional dimension in regard to ASD research which is the study of structural variation analysis in ASD. The DECIPHER database hosted at the Sanger center for genome sequencing, another genetic resource, provides additional information about the CNV loci associated with ASD. It’s a web-based interactive database which Wellcome Trust Sanger Institute maintains. More than 1,100 entries are available when the DECIPHER database is searched with the keyword “Autism”. The database can be accessed at http://decipher.sanger.ac.uk/.
Conclusion
Open-source bioinformatics resources can be valuable tools for biological research, providing access to a wealth of information and computational power. Before starting any project, it is important to identify the appropriate bioinformatics resources for your specific research question. Open-source bioinformatics resources are constantly evolving, and new updates are frequently released. A FAIR (Findability, Accessibility, Interoperability, and Reuse) approach to the databases is also helping researchers access the data more efficiently. The availability of open-access bioinformatics data repositories ranging from all-encompassing NCBI, EBI to specialized ASD databases and genomic resources has been a game changer for researchers who now can leverage bioinformatics data and tools to perform research on a simple laptop.
If you’re interested in open-source bioinformatics data repositories design, custom enhancement, research, and development, contact us by email at info@zifornd.com, and we can discuss further.