Evolution of the ACMG/AMP variant interpretation guidelines
Establishing the pathogenicity of variants identified in genes is at the heart of clinical genetic testing. In 2015, the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) released a set of variant interpretation guidelines that quickly became the industry standard. Over the years, groups like ClinGen have refined these guidelines and are working with a consortium on new standards for interpretation (v4), expected to be published this year or next. Concurrently, databases and other resources used in the application of these guidelines to make inferences about variants have grown and matured.
One of the simplest, yet most powerful indicators that a variant may be benign is the frequency with which it appears in the general, ostensibly healthy population. As many Mendelian disease genes that are targets of genetic testing have high penetrance, the expectation is that pathogenic variants in these genes should be at a low prevalence in the healthy population. Conversely, having a high allele frequency is used as strong evidence toward benign variant classification.
The ACMG 2015 guidelines included three allele frequency criteria. Over the years, more nuance was added to each of these criteria, as outlined in the table below.
| Criterion | Strength | Definition / Revisions |
|---|---|---|
| BA1 | Stand alone benign | >5% in ExAC, 1K Genomes or EVS databases Gene-specific cutoffs, Use of subpopulation frequencies |
| BS1 | Strong benign | Frequency higher than expected for the disorder Use of subpopulation frequencies |
| PM2 | Moderate pathogenic | Absent or at an extremely low frequency in controls Downgraded strength to supporting |
It is expected that a new version of the ACMG variant interpretation guidelines (v4) will be released in 2024/2025. A preview of those guidelines describes a further evolution of the allele frequency criteria. Central to this is the establishment of a disease allele frequency (DAF) that users define for each gene.
One common method to derive DAFs is from population allele frequencies of documented pathogenic variants. The idea is that, with so many genomes sequenced, the most common pathogenic variant in a population will have already been observed and that subsequent pathogenic variants will have a lower frequency.
During variant interpretation, the allele frequency of the variant is assessed against the gene’s DAF to see if any sub-population has a frequency above the DAF threshold. If so, it would support a benign classification. Falling below this threshold will no longer be used as evidence in support of pathogenicity (i.e., PM2 no longer holds weight).
Importantly, the new guidelines also make it easier to reach a classification of Likely Benign. Instead of requiring two pieces of evidence, a single piece of evidence, such as surpassing the DAF, can result in a classification of Likely Benign. This highlights the importance of getting the DAF threshold right.
gnomAD database
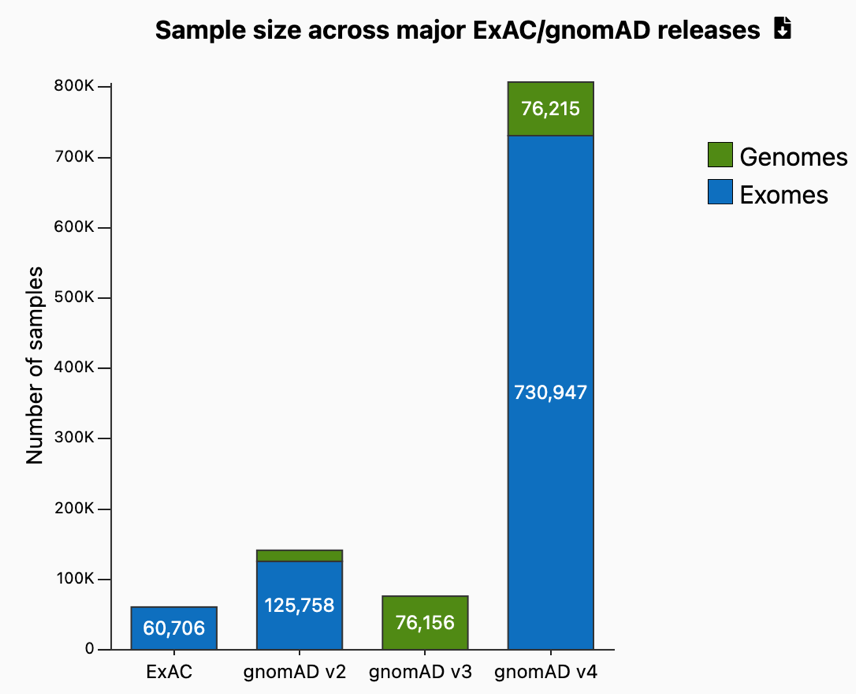
A decade ago, there were a few relatively small databases of sequence variants in healthy individuals: ExAC, 1K Genomes, EVS. Over time, the Broad Institute’s ExAC database grew and in 2017, they released gnomAD, the largest publicly available database of exomes and genomes from healthy individuals, along with a useful browser. gnomAD V2.1, released a few months later, remains the largest such dataset of variants aligned to the GRCh37 reference genome. gnomAD has since released additional versions of its dataset (V3 and V4) that contain a larger number of individuals from more diverse populations and aligned to the GRCh38 reference genome. gnomAD V4, released in 2023, contains over 800,000 genomes/exomes representing several continental sub-populations and additional diverse ancestry groups.
gnomAD presents global allele frequencies for each variant across the entire gnomAD database. In addition, to facilitate the identification of continental sub-populations with the highest allele frequency, gnomAD displays a new metric for each variant called the Grpmax FAF (this replaces PopMax). It is defined as the maximum credible genetic ancestry group allele frequency, calculated as the lower 95% confidence interval of that population’s allele frequency in gnomAD. If the Grpmax FAF for a variant is above the gene-specific DAF, then it supports the variant being benign. The strength of that support depends on how far above the gene-specific FAF the variant Grpmax FAF falls.
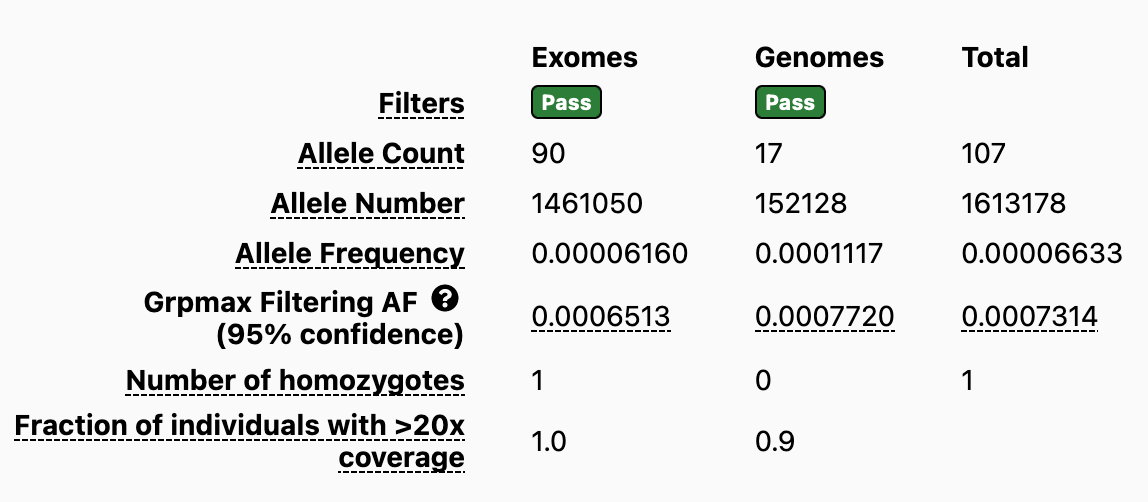
Impact of migrating to gnomAD V4 for labs
As labs consider migrating over to gnomAD V4, one pertinent question is whether thresholds set for DAFs and/or BA1/BS2/PM2 need to be revisited. Using DAF thresholds derived from gnomAD V2 and then using gnomAD V4 during variant interpretation could lead to unexpected results.
Fortunately, a review of a handful of CDC Tier 1 genes (table below) found that the Grpmax frequencies are typically less than an order of magnitude different between gnomAD V2 and V4, suggesting there may be little impact. Nonetheless, labs may want to review their BA1/BS2/PM2 thresholds prior to migrating to gnomAD V4 to ensure accurate results.
Highest pathogenic variant allele frequency in gnomAD for select genes (used to calculate DAF and BA1/BS2/PM2 thresholds)
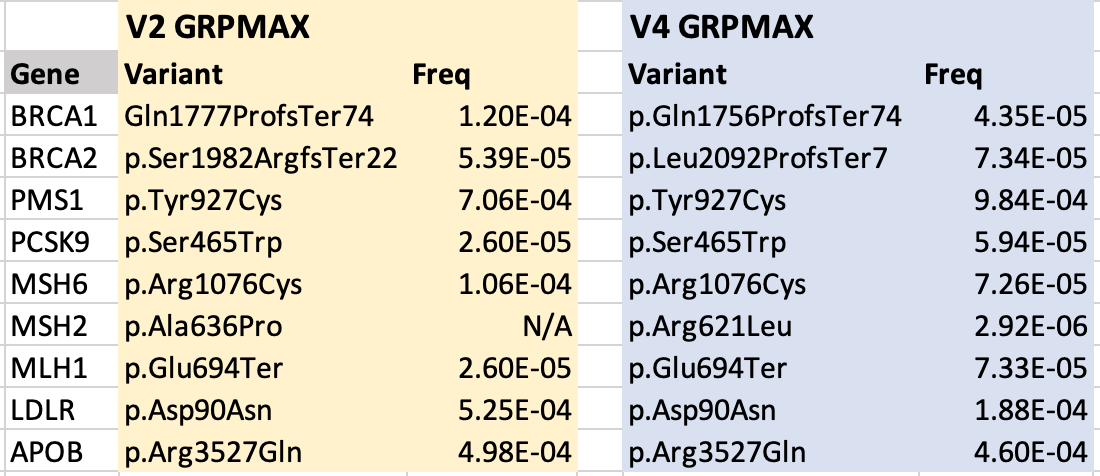
The availability of the new Grpmax FAF in gnomAD will make the tasks of both establishing allele frequency thresholds for a given gene and determining whether a given variant surpasses these thresholds much easier.
If you have any questions or if you would like to know more, please contact us at info@zifornd.com.






