Introduction
RNA and protein folding can have significant drug discovery and development application as they aid in understanding molecular structures, dynamics, and interactions.
In a drug discovery context, RNA sequence analysis tools can aid in identifying potential drug targets or regulatory elements in disease-related RNA sequences. The MicroRNA target prediction tool from ViennaRNA helps in understanding post-transcription regulation, which can be relevant in diseases such as cancer, where microRNA dysregulation is common.
Protein folding tools are crucial in understanding the three-dimensional structure of proteins, which are in turn essential for rational drug design. By studying the folding pathways and dynamics of target proteins, researchers can identify potential binding sites and design drugs that specifically interact with disease-related proteins. There are several large-scale computational projects such as Rosetta@Home, Folding@Home, Foldit which are focusing on protein folding. In addition the application of Artificial Intelligence (AI) on computational problems has resulted in the development of AlphaFold, by DeepMind & EMBL, which performs protein structure predictions using deep learning. This whole area will be a topic in a future blog
Protein folding tools can also help guide site-directed mutagenesis experiments to engineer proteins with desired properties. This has applications in biotechnology, such as the development of enzymes with enhanced catalytic activity or increased stability. In infectious diseases, understanding the three-dimensional structure of viral or bacterial proteins is crucial for designing effective vaccines. In this case, protein folding tools can assist in predicting antigenic regions and developing vaccines that elicit a robust immune response.
Here's a workflow that combines ViennaRNA and protein folding tools to predict the secondary structure of an RNA sequence followed by modelling the 3D structure of the associated protein.
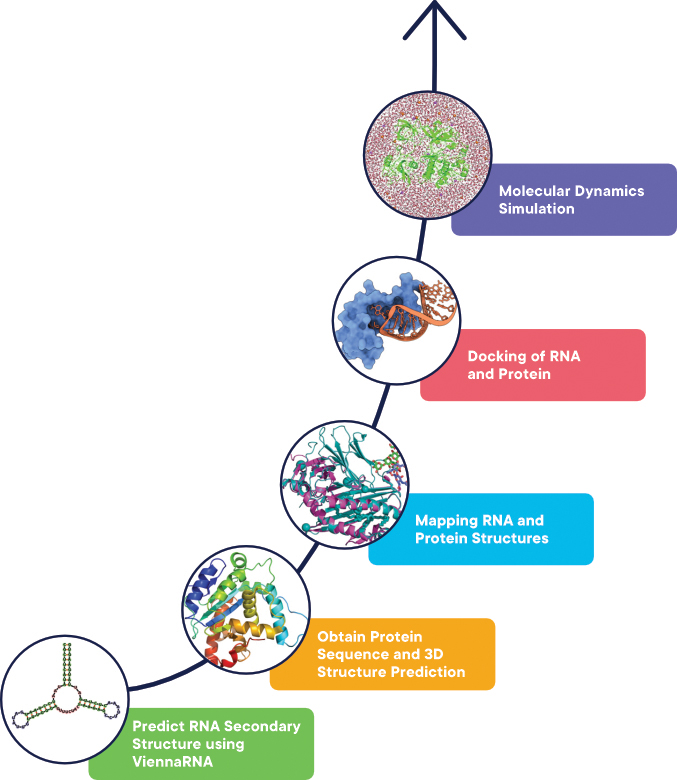
Figure 1. A simple workflow which combines RNA and protein sequences tools for modelling and analysis.
Below is the description of the tools listed in the workflow (Figure 1) with each step describing the tools required, the URL of the tools and a high-level overview of executing the tool.
Step 1: Predict RNA Secondary Structure using ViennaRNA
- Install ViennaRNA: Download and install ViennaRNA package from the official website (https://www.tbi.univie.ac.at/RNA/) or use the appropriate package manager.
- Prepare RNA Sequence: Obtain the RNA sequence you want to analyze. Save it in a plain text file, e.g., "rna_sequence.txt". The format should be in a FASTA file format.
- Run RNAfold: Use the ViennaRNA "RNAfold" command to predict the secondary structure of the RNA sequence. Open a terminal and execute the following command:
“RNAfold < rna_sequence.txt > rna_structure.txt”.
This command will output the predicted secondary structure in "rna_structure.txt" along with the calculated minimum free energy (MFE) and ensemble free energy.”
Step 2: Obtain Protein Sequence and 3D Structure Prediction
- Obtain Protein Sequence: If you don't have the protein sequence already, you can obtain it from publicly available databases or predict it using tools like BLAST or PSI-BLAST. The NCBI is the best place to learn more about downloading and installing BLAST suite of tools.
- Predict Protein 3D Structure: Use a protein structure prediction tool such as I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) or Rosetta (https://www.rosettacommons.org/software) or SWISS-MODEL (https://swissmodel.expasy.org/) to predict the 3D structure of the protein based on its sequence.
Step 3: Mapping RNA and Protein Structures
- Identify RNA-Protein Interactions: Use RNA-protein interaction prediction tools like RPISeq (http://pridb.gdcb.iastate.edu/RPISeq/) or RPI-SE ( https://github.com/haichengyi/RPI-SE/) to identify potential RNA-binding sites on the protein.
- Visualization: Use molecular visualization software like PyMOL (https://pymol.org/) or ChimeraX (https://www.cgl.ucsf.edu/chimerax/) or VMD (https://www.ks.uiuc.edu/Research/vmd/) to visualize both the predicted RNA secondary structure and the modeled protein 3D structure. You can superimpose the structures to analyze potential RNA-protein interactions.
Optional steps
Docking of RNA and Protein
- Docking Prediction: Use a molecular docking tool like ClusPro (https://cluspro.bu.edu/) or HADDOCK (https://www.bonvinlab.org/software/haddock2.4/) to perform the docking of the RNA secondary structure (from Step 1) with the protein structure (from Step 2).
- Analyze Docking Results: Analyze the output of the docking simulation to identify potential RNA-protein binding sites and evaluate the interactions.
Molecular Dynamics Simulation
If you want to study the dynamics of the RNA-protein complex, you can perform molecular dynamics (MD) simulations using tools like GROMACS (https://www.gromacs.org/) or AMBER (https://ambermd.org/).
RNA folding tools are crucial for predicting the secondary structure of RNA, which is essential for understanding their function and interactions. To achieve accurate results, data preprocessing is a critical step. Some important considerations in data preprocessing for RNA folding experiments are: sequence format conversion, sequence length Normalization, identification & handling of pseudoknots, data augmentation, secondary structure parsing and large dataset handling.
Downstream analysis of RNA folding results involves the examination and interpretation of the predicted RNA secondary structure obtained from RNA folding algorithms. These predictions are typically represented in various formats, such as dot-bracket notation, which describes the base-pairing interactions in the RNA molecule.
The downstream analysis of RNA folding results is a multi-faceted process that combines computational predictions with experimental data and functional annotations to gain a deeper understanding of the RNA's structure and function. There are several factors such as identifying structural motifs, energy analysis, conservation analysis, interaction prediction, functional analysis and visualization which all combine to provide a deep understanding of the results generated.
Remember that predicting protein 3D structures is a challenging task and may not always be accurate. Additionally, the reliability of RNA-protein interaction predictions can vary depending on the tools and parameters used. Therefore, it's essential to interpret the results with caution and validate the findings through experimental techniques if possible.
If you're interested in RNA & Protein folding & sequence analysis or other bioinformatics specialties, please contact us at info@zifornd.com