The relentless pursuit of new therapeutic targets fuels the engine of drug discovery. Enter Open Targets, a robust platform that harmonizes the vast array of bioinformatics data sources to systematically identify and prioritize promising targets for a wide range of diseases. This article delves into the intricate workings of Open Targets, exploring how it integrates a multitude of bioinformatic data sources and translates them into a powerful scoring system for target-disease associations.
A Complex Tapestry of Bioinformatics Data Sources
Open Targets draws upon a rich tapestry of publicly available bioinformatics data, each thread offering a unique perspective on the biological landscape of disease. Here’s a glimpse into some key sources that contribute to the platform:
- Genetic Whispers: Genome-wide association studies (GWAS) reveal genetic variations linked to disease susceptibility. Open Targets listens to these whispers, identifying genes harboring these variations—potential targets for therapeutic intervention.
- Somatic Mutations: Mutations within diseased tissues can expose genes critical for disease. The platform integrates data on somatic mutations from cancer sequencing projects, providing valuable insights.
- The Drug-Target Tango: Existing drugs and their established targets offer a wealth of knowledge. Open Targets incorporates information on approved drugs and their targets to help identify potential targets for similar diseases, facilitating a targeted drug discovery approach.
- Differential Expression’s Crescendo: Gene expression profiling across healthy and diseased tissues can reveal genes with altered activity. Open Targets integrates transcriptomics data to identify genes whose expression levels rise or fall in disease states, potentially offering new therapeutic insights.
- Animal Models: Understanding the biological pathways and networks associated with diseases can illuminate potential targets. The platform integrates data on pathways and interactions between genes and proteins, revealing insights into the intricacies of disease.
- Pathway Harmony: Understanding the biological pathways and networks associated with diseases can illuminate potential targets. The platform integrates data on pathways and interactions between genes and proteins, revealing insights into the intricacies of disease.
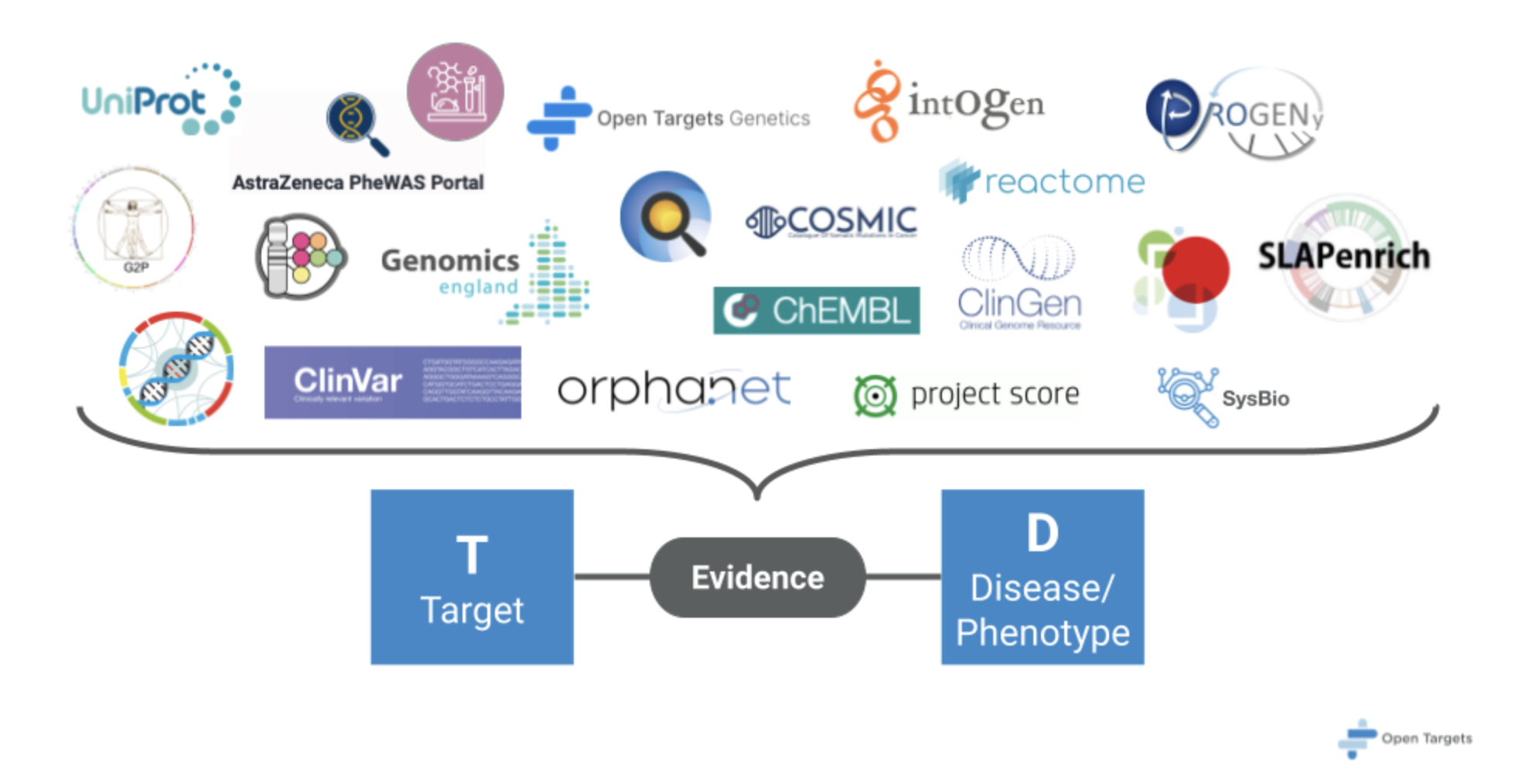
Figure 1. Heterogenous data sources form the backbone of the evidence required for target disease associations.
Release 24.06 of Open Targets has the following statistics:
- 63,226 targets
- 28,198 diseases and phenotypes
- 18,041 drugs and compounds
- 17,703,456 evidence strings
- 8,079,215 target-disease associationsThis release integrates 17,703,456 evidence strings to build 8,079,215 target-disease associations between 28,198 diseases and 63,226 targets from the following 23 public resources. Additionally, the platform now allows users to explore data on 18,041 drugs.
Table 1: List of evidence sources for the Open Targets platform.
Open Targets Evidence Summary
| Data Source | Category | Number of Entries | Reference |
|---|---|---|---|
| European Variation Archive (EVA) | Genetic | 3,260,235 | [2] |
| Open Targets Genetics | Genetic | 781,213 | [3] |
| Gene2Phenotype | Genetic | 3,631 | [1] |
| Genomics England PanelApp | Genetic | 34,407 | |
| ClinGen | Genetic | 2,642 | |
| Orphanet | Genetic | 6,673 | |
| Gene Burden | Genetic | 36,372 | |
| CRISPRBrain | Genetic | 21,726 | |
| UniProt Literature | Genetic | 4,505 | [4] |
| European Variation Archive (EVA) | Somatic | 16,962 | [2] |
| intOGen | Somatic | 4,359 | |
| Cancer Gene Census | Somatic | 82,754 | |
| Uniprot | Somatic | 28,354 | [4] |
| ChEMBL | Drug Target | 660,575 | [1] |
| Expression Atlas | Gene Expression | 230,182 | |
| Reactome | Pathway | 10,181 | |
| SLAPenrich | Pathway | 72,441 | |
| PROGENy | Pathway | 378 | |
| SysBio | Systems Biology | 389 | |
| Cancer Genome Interpreter | Somatic | 1,300 | |
| Pacini et al. (2024) | CRISPR-Cas9 (Cancer Cell Lines) | 517 | [5] |
| IMPC | Mouse Model | 1,138,754 | |
| Europe PMC | Literature Co-occurrence | 11,304,906 |
Note:
- Numbers are based on the information provided and may not represent the exact number of entries in each data source.
- References are provided where available. 1: denotes reference not provided in snippet, 2: [2] refers to European Variation Archive (EVA), 3: [3] refers to Open Targets Genetics, 4: [4] refers to UniProt Literature, 5: [5] refers to Pacini et al. (2024).
Bioinformatics Workflows: The Unsung Conductor
Managing and analyzing the plethora of data utilized by Open Targets necessitates robust bioinformatics workflows and pipelines. These automated, multi-step processes ensure efficient and reproducible data processing. Here’s a simplified breakdown of a typical workflow conducting Open Targets’ bioinformatics orchestra:
- Data Acquisition: Scripts and data tools need to be nimble, automatically retrieving data from various public repositories.
- Data Preprocessing: Downloaded data often requires cleaning and standardization to ensure compatibility with downstream analyses. Workflows perform tasks like filtering irrelevant data points or converting data formats.
- Data Integration: Data from diverse sources needs to be integrated into a cohesive knowledge base. Workflows use mapping strategies to link entities (genes, diseases, drugs) across different datasets.
- Data Analysis: Depending on the data type, workflows execute specific bioinformatics tools, such as RNA-seq data analysis or GWAS data analysis, employing statistical rigor to identify significant associations.
- Target-Disease Association Scoring: Workflows aggregate the results from individual analyses and calculate the final score for each target-disease association, creating the final all-important insights.
Computational Resources
Unfortunately, Open Targets isn’t designed to be easily installed and run as a local platform. It’s a complex system leveraging significant computational resources. Having said that, the Open Targets team has released a tool on GitHub called “Standalone local deployment for Open Targets Platform”, which uses the GNU make command to install a local version of the Open Targets platform. Below is the architecture diagram for the local installation:
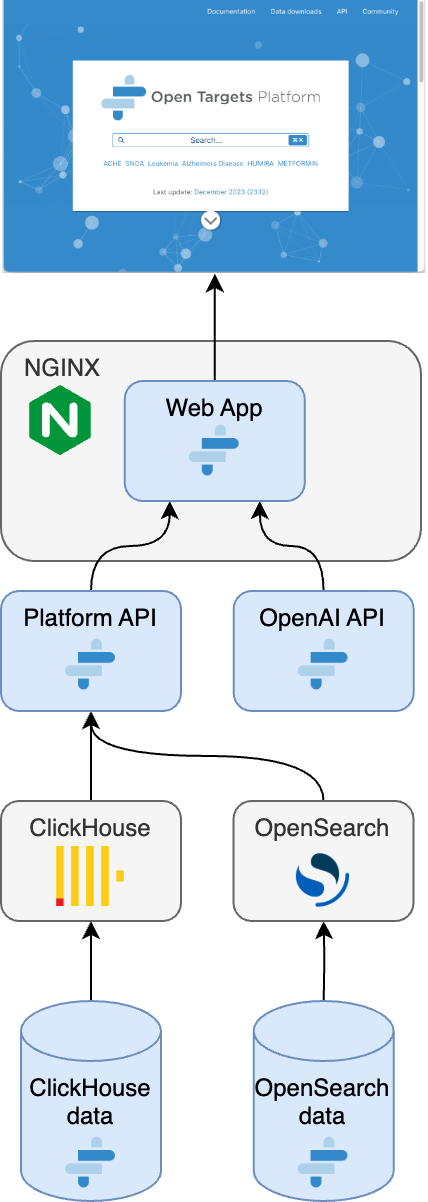
Figure2. Overview of the stand-alone installation of the Open Targets platform.Source:
https://github.com/opentargets/standalone-deployment-platform?ref=blog.opentargets.org
There are some challenges when trying to run a local instance of Open Targets:
- Large-scale data storage: Open Targets integrates a vast amount of data from diverse sources. This data would require significant storage (potentially terabytes) depending on the specific data selected, which is not typically possible on a desktop PC.
- Computational power: The platform utilizes bioinformatics workflows involving various tools and analyses. These workflows can be computationally intensive, requiring powerful processors and potentially graphics processing units (GPUs) for specific tasks.
- Software Dependencies: Open Targets likely relies on a complex software stack with various bioinformatics tools and libraries. Setting up these dependencies on a local machine can be challenging and time-consuming.
- Scalability and Maintenance: Open Targets is constantly evolving and integrating new data sources. Maintaining a local instance would necessitate manual updates and potentially significant reconfiguration efforts.
Beyond the extensive trove of public datasets, Open Targets also delves into the world of biomedical literature. A bioinformatics pipeline powered by BioBERT, a natural language processing (NLP) model specifically trained on biomedical text, is employed. BioBERT identifies mentions of genes, diseases and drugs within abstracts and full-text articles by using a method called NER(Named entity recognition) . NER is a form of natural language processing (NLP) that involves extracting and identifying essential information from text. By using more nuanced models like LinkBERT , ClinicalBERT and quite possible LLMs it maybe possible to enhance the pipeline’s ability to extract knowledge from these datasets. [see slides 8, 15,16 and 17 for reference if required]. This extracted information is then normalized using identifiers and synonyms to link it to entities within the platform’s knowledge base. Using more nuanced models like LinkBERT, ClinicalBERT employing NER (Named Entity Recognition) and possibly LLMs (Large Language Models) can enhance the pipeline’s ability to extract knowledge from these datasets.
Architecture and Overview of the Open Targets Platform
Typically, an Open Targets architecture leverages a combination of:
- Distributed data storage systems.
- Workflow managers for data processing pipelines.
- Bioinformatics tools for specific data analysis.
- Natural language processing for literature mining.
- A user-friendly web interface for data access and exploration.
Major components include:
- Data storage modules and components.
- Annotation modules.
- Evidence generation and target association.
- Web application and front-end.
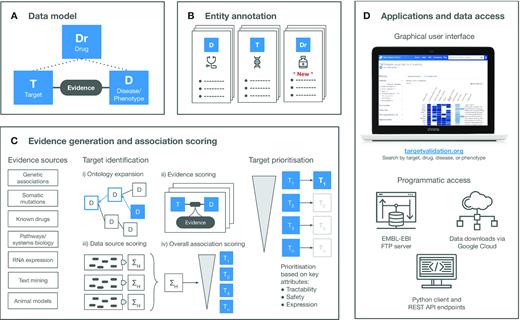
Figure 3. Overview of a typical architecture of the Open Targets platform.Source:
https://doi.org/10.1093/nar/gkaa1027
There are a number of critical challenges associated with the utilization of the Open Targets platform:
- Data Interpretation and Integration:
- Open Targets integrates a vast amount of data from diverse sources. This presents challenges in data interpretation. Users need a strong understanding of bioinformatics and the specific data types involved to properly evaluate the evidence supporting each target-disease association. Additionally, integrating data from various sources with potential inconsistencies or biases requires careful consideration.
- Data Quality and Completeness:
- Open Targets relies on publicly available data sources, which may have inherent limitations in quality and completeness.Users need to be critical of the data and consider factors like potential biases, missing data points, or methodological limitations of the original studies.
- Focus on Known Genes:
- Open Targets prioritizes established genes with existing annotations. This can lead to overlooking novel or less-studied genes with potential as drug targets. Researchers needing to explore entirely new avenues may need to complement Open Targets with other resources.
- Target Druggability:
- While Open Targets identifies potential target-disease associations, it doesn’t explicitly assess druggability. This is a crucial factor in drug discovery, as not all potential targets are readily amenable to therapeutic intervention. Additional resources or computational tools focusing on druggability prediction may be needed.
- Visualization:
- Platform Complexity: Open Targets offers a wealth of information, but the platform itself can be complex to navigate, particularly for users unfamiliar with bioinformatics concepts. A steep learning curve can be a barrier to entry, especially for researchers lacking a strong background in this area.
There are also a number of additional considerations:
- Computational Demands:
- While users aren’t required to run the platform itself, downloading and analyzing large datasets from Open Targets may require significant computational resources on their local machines.
- Data Updates:
- Open Targets is constantly evolving, integrating new data sources and refining scoring methods. Users need to stay updated on these changes to ensure they are utilizing the most current information.
In addition to the above lists, one of the key considerations is the scoring algorithm deployed by the platform. A brief overview of the target-disease association scoring is given below.
Building and Scoring Target-Disease Associations
The power of Open Targets lies in its ability to integrate diverse bioinformatics data sources and generate a comprehensive score for each target-disease association. This score reflects the cumulative evidence supporting a particular gene product (protein) as a potential target for a given disease. Here’s a simplified breakdown of the scoring process:
- Each data source contributes a specific weight to the overall score, reflecting its perceived importance in target validation.
- For instance, genetic associations from GWAS might hold a higher weight than findings from animal models.
- Within each data source, individual data points (e.g., a specific mutation or a differentially expressed gene) also contribute a score based on factors like statistical significance or fold-change in expression.
- The platform aggregates these individual scores from each data source, resulting in a final, cumulative score for the target-disease association.
Strategies for Mitigating Challenges:
- Critical Evaluation of Data: Each data source contributes a specific weight to the overall score, reflecting its perceived importance in target validation.
- Complementary Resources: For instance, genetic associations from GWAS might hold a higher weight than findings from animal models.
- Collaboration: Within each data source, individual data points (e.g., a specific mutation or a differentially expressed gene) also contribute a score based on factors like statistical significance or fold-change in expression.
- Staying Updated: The platform aggregates these individual scores from each data source, resulting in a final, cumulative score for the target-disease association.
Summary
The Open Targets platform is a comprehensive, open-source research tool that integrates publicly available datasets to support the systematic identification and prioritization of potential therapeutic drug targets. It scores target-disease associations by combining data from various sources, including genetics, pathways and chemical compounds. Researchers can use this platform to explore and analyze drug targets, aiding drug discovery and development efforts.
The Open Targets platform exemplifies the power of bioinformatics in drug discovery. By integrating and analyzing a vast array of bioinformatics data, Open Targets empowers researchers to prioritize promising drug targets with a strong biological rationale. This not only accelerates the drug discovery process but also increases the likelihood of identifying effective therapies for a multitude of diseases.
As bioinformatics methodologies continue to evolve, we can expect the Open Targets platform to become even more sophisticated, offering researchers an ever-more powerful tool in the fight against disease. In summary, Open Targets is a valuable resource for advancing precision medicine and improving patient outcomes.