Introduction
cBioPortal is an open-source platform that visualises, analyses and serves large-scale cancer genomics data sets. Ingesting data from cBioPortal into a custom database involves several steps and challenges that must be addressed to ensure data integrity and usability.
Data sources
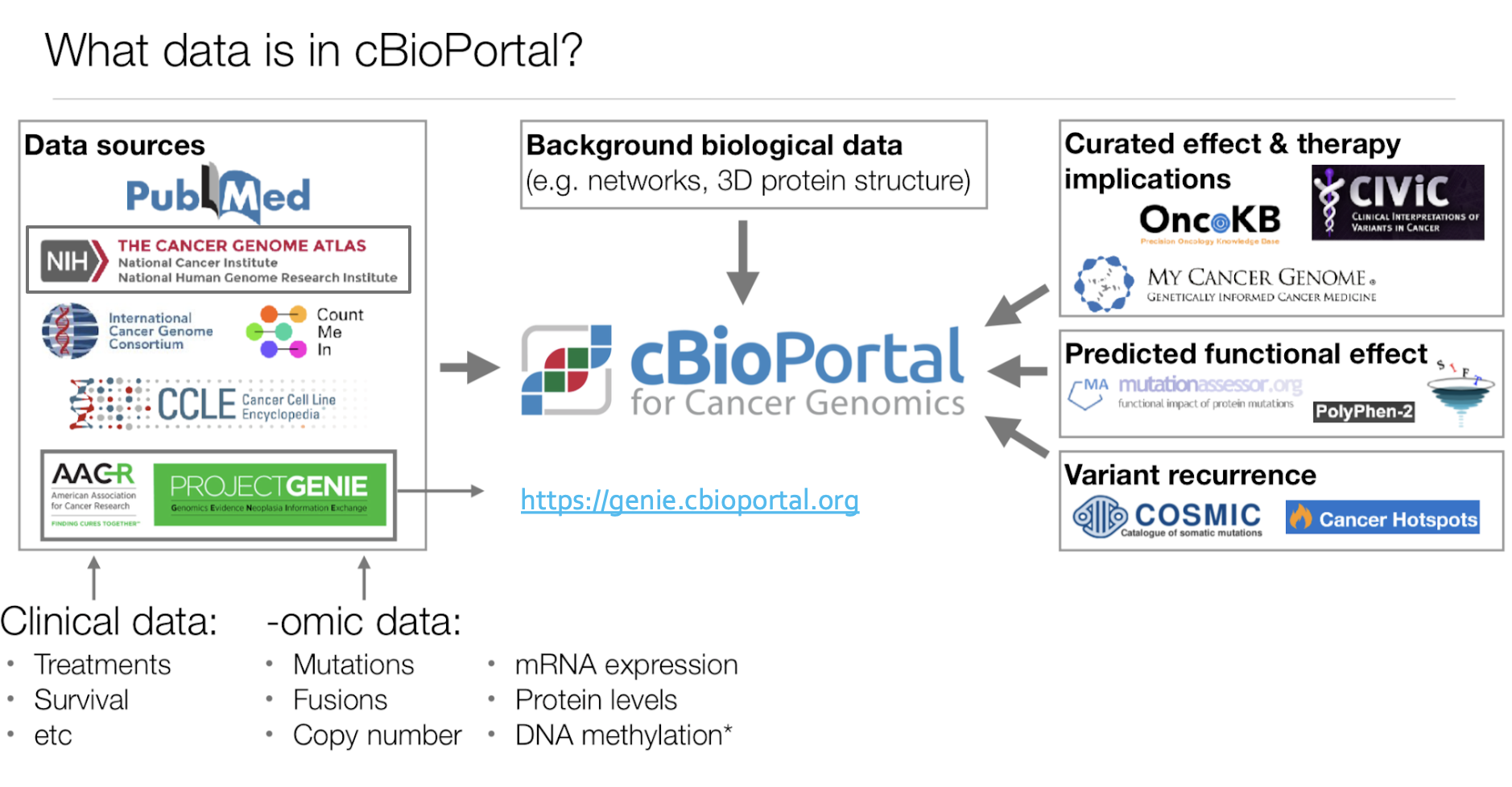
The frequency of data updates depends on the availability of new datasets and curation efforts. The Cancer Genome Atlas (TCGA) data within cBioPortal is continuously updated to reflect the latest findings. It includes genomic alterations, copy number variations, mutations, and clinical attributes. TCGA data in cBioPortal is comparable to TCGA data in the Genomic Data Commons (GDC), and some legacy data, such as TCGA Firehose data, are also available.
As of the latest update, cBioPortal has added 5,120 samples from 13 studies, highlighting the platform's extensive and growing database.
Table 1: cBioPortal latest Datasets update 2023
| Latest update Dec 29, 2023 | Name |
|---|---|
| Study Name | Samples |
| Endometrial Cancer (MSK, Cancer Discovery 2023) | 1,882 |
| Esophagogastric Cancer (MSK, J Natl Cancer Inst 2023) | 902 |
| Diffuse Glioma (GLASS Consortium, 2022) | 693 |
| Bladder Cancer (MSK, Clin Cancer Res 2023) | 526 |
| Non-Small Cell Lung Cancer Brain Metastasis (MSK, Nat Commun 2023) | 322 |
| MSK Make-an-IMPACT Rare Cancers (MSK, Clin Cancer Res 2023) | 184 |
| Cervical Cancer (MSK, Clin Cancer Res 2023) | 177 |
| Mature T and NK Neoplasms (MSK, Blood Adv 2023) | 132 |
| Hepatocellular Carcinoma (MSK, JCO Precis Oncol 2023) | 90 |
| Sarcoma (MSK, J Pathol 2023) | 82 |
| Esophagogastric Cancer (MSK, Clin Cancer Res 2023) | 64 |
| Rhabdomyosarcomas (MSK, NPJ Precis Oncol 2023) | 42 |
| Pediatric Rhabdomyosarcomas (MSK, JCO Precis Oncol 2023) | 24 |
cBioPortal Computational requirements
| Platform | Instance type | (v)CPUs | RAM(GB) | Storage (GB) |
|---|---|---|---|---|
| aws | r5.xlarge | 4 | 32 | 50 |
| on-premise | - | 4 | 32 | 50 |
A minimal computational infrastructure is required to run the cBioPortal instance, depending on the number of daily users and data loaded in the portal.
Installation and Data download
Local installation of cBioPortal can be done using Docker, which will then deploy and start all 4 containers listed below:
- Mysql database, which holds all the portal's data.
- cBioPortal Java web app, which serves as the REACT frontend & REST API.
- Sessions Service Java web app, which stores session information.
- Mongo database that persists the data for the sessions service.
Challenges
- Maintaining updates and modifications requires running these containers in a different cloud or on-premise computational ecosystem.
- Sometimes, the Mongo database does not connect from a local instance and times out, which requires recalibration and fine-tuning of computational resources.
Below is the latest overview of the datasets available in cBioPortal for public review.
Most of the CBioPortal data is available only in TSV format, which might create a challenge in the downstream or tertiary analysis if a different format such as VCF or MAF, is required.
Table 1: cBioPortal Datasets Overview
| Datasets available | 403 |
| Mutations | 182,880 |
| CAN | 138,689 |
| RNA-Seq | 31,179 |
| Structural Variants | 33,336 |
| Tumor mRNA (microarray) | 8,506 |
| Tumor miRNA | 2,882 |
| Methylation (HM27) | 4,564 |
| RPPA | 18,792 |
| Protein Mass Spectrometry | 1,687 |
| Complete | 27,737 |
| Treatment Count | 8,612 |
cBioPortal Architecture – key components
- Backend - SQL database
- Validator - checking format before importing data into the database
- Frontend – built with React, Mobx & Bootstrap
- Session Service – Rest API & Mongo Database
- API from external sources*
- Multiple API clients in R & Python are available; the most recommended are the Bioconductor package (cBioPortalData) & cBioPortalR.
- OncoKB - Precision oncology knowledgebase – effects & treatments of specific cancer gene alterations.
- CIVIC - Clinical Interpretation of Variants in Cancer
- GenomeNexus – High-throughput annotation of variants in cancer
- G2S – Genome to Structure – maps variants to 3D structure
- CancerHotSpots – Linking statistically significant mutations in cancer is determined using a binomial statistical model and is not experimentally validated or curated by a literature review.
- Multiple API clients in R & Python are available; the most recommended are the Bioconductor package (cBioPortalData) & cBioPortalR.
Challenges in Data Ingestion from cBioPortal
- Data Complexity: cBioPortal's data encompasses various types, including genomic, clinical, and molecular data. The complexity requires careful handling to maintain the relationships and integrity during Ingestion and migration.
- Data Volume: The large volume of data in cBioPortal can be challenging to manage, requiring scaled-up storage and processing capabilities.
- Data Consistency: Maintaining data consistency during transfer is crucial to prevent analysis errors.
- Data Compatibility: Custom & external databases may have different data structures, necessitating transformation or mapping of cBioPortal data to fit these structures.
- Identifier Mapping: Accurate mapping of identifiers like Entrez Gene IDs and HUGO Gene Symbols is essential for referential integrity.
- API Limitations: The cBioPortal API may have request limitations, affecting data transfer efficiency. Listed above* are a few external data sources requiring separate access protocols.
- Data Privacy: Ensuring the privacy of patient data is paramount, which may require additional security measures.
Gene-Specific Identifiers in cBioPortal
cBioPortal uses standardised identifiers to ensure consistency and accuracy in data representation and querying:
- Entrez Gene IDs Numeric identifiers assigned by the NCBI to genes, widely used for genomic data analysis.
- HUGO Gene Symbols: Standardised symbols provided by the HGNC serve as a common language for the genetics community.
These identifiers are essential for accurately querying and analysing genomic data within cBioPortal and when transferring data to other databases.
Additional Challenges
After careful analysis and review of the cBioPortal data structures, system architecture, analytics capabilities and overall entity relationships, below are some additional challenges and bottlenecks users will face when trying to ingest/migrate custom data into or out of cBioPortal.
- Some missing metadata & study data which might not be integrated into the MultiAssayExperiment object in the cBioPortal.
- Need to clear Cache when the download is interrupted and no proper way to check the integrity of the download.
- Use the "removeCache " function. Note that this function only works for data downloaded through the "cBioDataPack" function.
- Prior Z-Score Data normalisation & transformation of the input file is necessary before uploading it to the portal.
- Two recommended methods available
- ConvertExpressionZscores.pl (diploid samples as base population)
- NormalizeExpressionLevels_allsampleref.py (all samples as base population).
- Two recommended methods available
- Multiple API errors for the external data sources listed in the Github issues page.
- Data download issues were recently reported for the seed database.
- No clear instructions on downloading mutations/altered samples of whole genes in one clinical study as the current version can only download the data of mutations/altered samples in serval genes.
- Annotation of mutation data and transcripts needs to follow specific guidelines alternating between MKSCC & UNIPROT database for different sets of data.
- It is very important to annotate the mutation data in the same way else the gene + protein changes would be different when comparing 2 studies.
- Validation of altered samples in a study for all the genes is a challenging and a time-consuming process.
- ABCA7 gene has 10 altered samples in the main query page, whereas in the DNA_mutation.txt file it has listed only 7 altered samples.
- Structural Variants (SV) data is experimental and needs validation and can also be linked to external sources such as dbVar (https://www.ncbi.nlm.nih.gov/dbvar/content/human_hub/) which requires customised API's and connections to various data sources.
- Pathway analysis and download feature is not available and graph data analysis for overlapping, similar and p-value correlated pathways are not available.
- Cytoscape plugin and the CyNDEX-2 app is required to view pathways features like network analysis.
Ingesting data from cBioPortal to a custom database (either in the cloud or on-premise) is a complex process that requires careful planning and execution. A deep understanding of the portal's data sources, correlation of data entities, portal architecture, computational expertise, addressing the portal's challenges and developing solutions are crucial for successful data transfer and utilisation.
Some approaches for resolving missing metadata in cBioPortal are to inspect Data files and descriptions and check for any discrepancies in sample or patient IDs across different studies. In addition to these recommendations, the FAIRification of data generated from cBioPortal would also help researchers effectively analyse cancer Omics data. Aspects of Machine Learning (ML) applications in cBioPortal analytics will significantly impact areas such as Cancer subtype classification, Data integration and visualisation.
If you're interested in open-source bioinformatics data repository design, custom enhancement, data migration/Ingestion, research and development, contact us by email info@zifornd.com, and we can discuss further.


