Spatial transcriptomics is revolutionizing our understanding of gene expression patterns and disease states. However, harnessing the full potential of this technique requires careful consideration of a few critical elements, such as input data types and file formats, computational challenges, and the all-important downstream visualization.
Input Data: A Diverse Spectrum of Data Types and Formats
Spatial transcriptomics leverages a wide range of data types, each with unique characteristics.
Common types include:
- Spatial RNA sequencing (spRNA-seq): Generates gene expression count matrices for individual cells or spots within a tissue.
- In situ sequencing (ISS): Produces high-resolution images of gene expression patterns within cells.
- Slide-seq: Captures gene expression profiles on a spatially barcoded slide.
Understanding the specific data type and characteristics of your data is crucial for selecting appropriate analysis tools and methods, requiring the following steps:
- Data preprocessing: Cleaning, filtering, and normalizing the data to remove noise and artifacts.
- Spatial domain identification: Identifying biologically meaningful spatial regions or compartments within the tissue.
- Cell type annotation: Assigning cell types to different spatial locations based on gene expression profiles.
- Cell-cell interaction analysis: Inferring interactions between cells based on their spatial proximity and gene expression patterns.
Table 1 lists the files which are commonly available or required as input to a spatial transcriptomics analysis pipeline.
| Type | Name | Description |
|---|---|---|
| H5 File | raw_feature_bc_matrix.h5 | Contains raw feature-barcode matrix |
| JSON File | scalefactors_json.json | Contains scale factors for image alignment |
| PNG Image | tissue_hires_image.png | High-resolution tissue image |
| tissue_lowres_image.png | Low-resolution tissue image | |
| CSV File | tissue_positions_list.csv | List of tissue positions |
| gene_graph_clusters.csv | Clustering results from SpaceRanger | |
| parameters.csv | Metadata about the experiment | |
| TIFF Image | wsi.tif | High-resolution whole slide image |
| GeoJSON File | annotations.geojson (optional) | Defines regions of interest |
| HTML File | web_summary | Summary of the experiment |
Table 1: Commonly available input files, format and description utilized by spatial transcription pipelines.
Computational Challenges: A Complex Maze
Analyzing spatial transcriptomics data presents significant computational challenges due to the large volume and complexity of the data. Key challenges include the need for specialized computational tools and pipelines, which are essential for effective data processing.
A critical computational requirement is the availability of High-Performance Computing (HPC) infrastructure with a combination of GPUs + CPUs (Figure 1). Additionally, sufficient RAM and storage (cloud-based S3 or on-premise server storage) are needed for efficient execution of any spatial transcriptomics pipeline.
In addition, other key considerations for the computational requirements of Spatial Omics include:
- Data storage and processing: ST datasets can be large, requiring significant computational resources for storage and analysis. High-performance computing infrastructure may be necessary for efficient processing and analysis.
- Algorithm complexity: Many ST analysis algorithms are computationally intensive, especially for large datasets or complex spatial patterns. Developing efficient algorithms is crucial for practical applications.
- Docker networking & API interface (plumber):
- Plumber API deployed in different containers.
- Interaction between containers via sending request and receiving response.
- Python packages (Hydra and Metaflow):
- Metaflow was used to build and design the pipeline workflow.
- Hydra used to generate dynamic configuration capabilities.
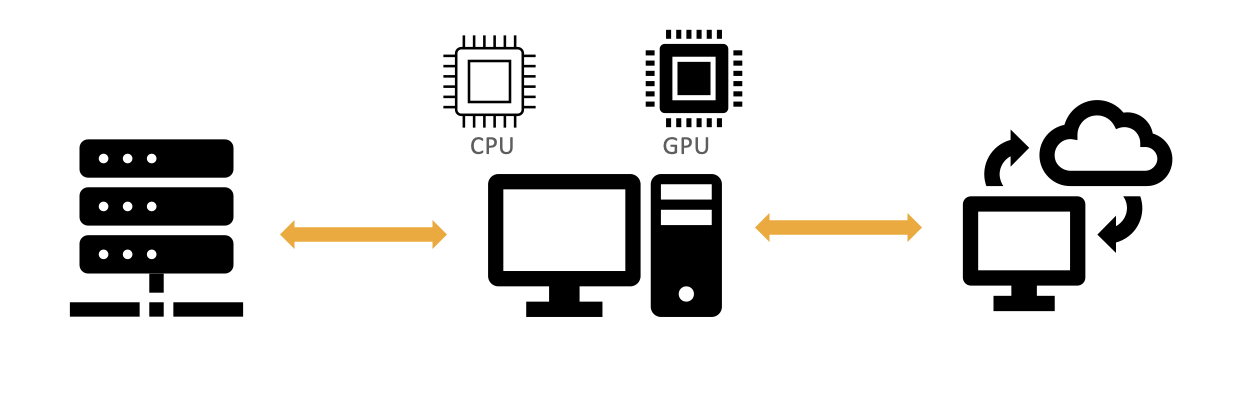
Figure 1: A typical high performance computational (HPC) system with GPU + CPU for executing spatial transcriptomics pipelines.
Downstream Visualization: A Clear Picture
Visualization of spatial transcriptomics results is critical for understanding gene expression patterns within a tissue. Researchers can identify spatial domains, cell types, and gene-gene interactions by combining spatial information with gene expression data. This enables a deeper understanding of biological processes and disease mechanisms.
Creating informative and visually appealing representations of spatial gene expression patterns is essential. Visualizing spatial transcriptomics data is crucial for understanding and communicating biological insights. Effective visualization techniques include:
- Heatmaps: Representing gene expression levels across different cells or spatial regions.
- Spatial plots: Displaying gene expression patterns within the tissue context.
- 3D visualizations: Creating three-dimensional representations of spatial gene expression data.
- Interactive tools: Allowing users to explore and analyze data interactively.
Choosing the right visualization methods depends on specific research questions and the nature of the data.
Figure 2 shows a typical visualization of the spatial data results using Tissuumaps, an open-source tool that helps view different aspects of the results.
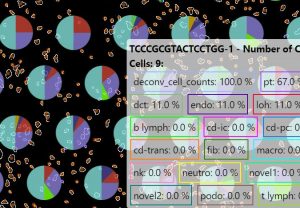
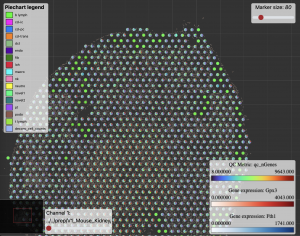
Figure 2: Spatial transcriptomics visualization of results used for downstream analysis using Tissuumaps.
In addition to this open-source tool, custom visualization options require developing R Shiny applications that provide data visualization spanning initial data processing, cell clustering, annotation and UMAP plots for deeper insights. Figure 3 shows a custom R Shiny app displaying UMAP data, ready for analysis.
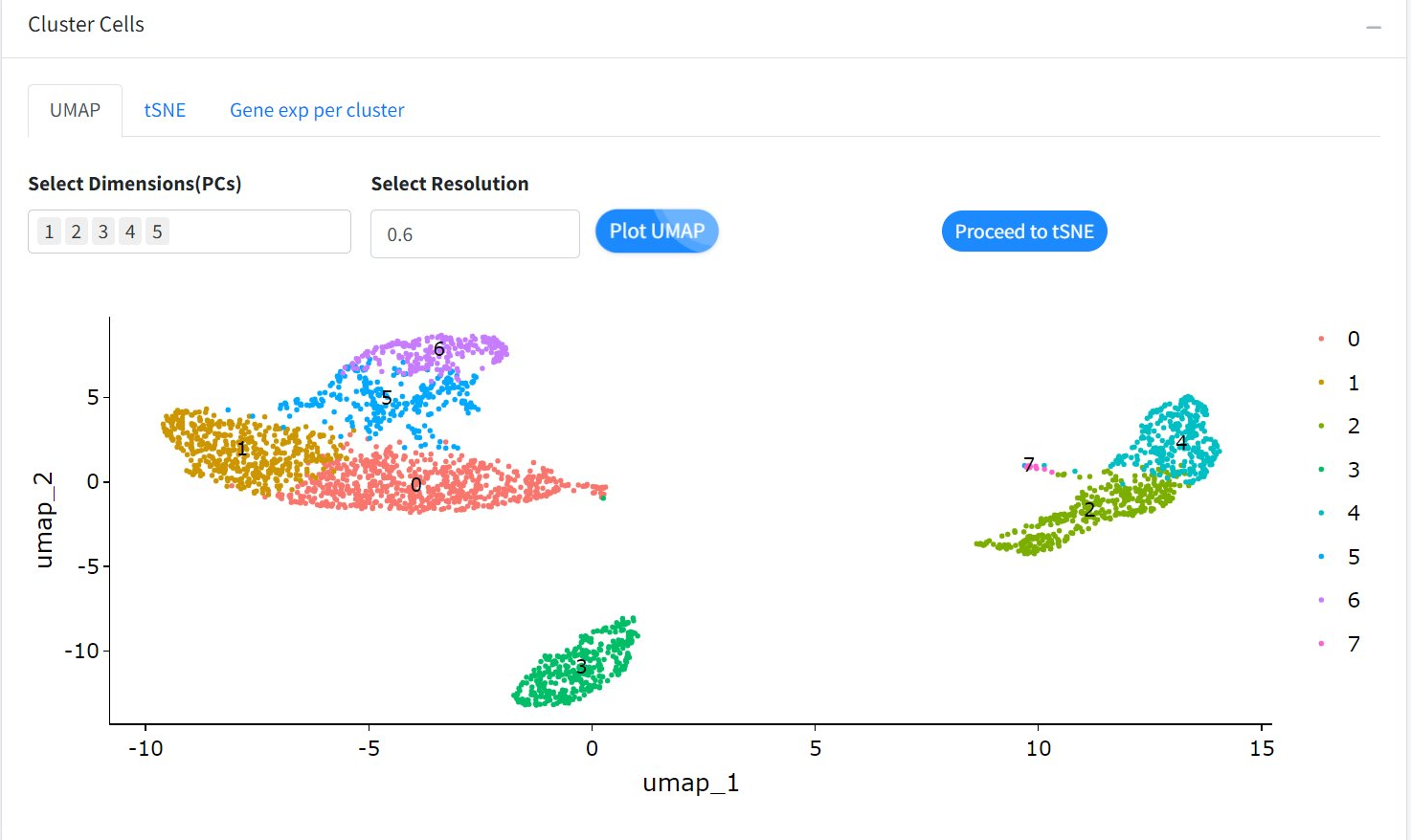
Figure 3: Custom R Shiny app depicting spatial omics data ready for analysis.
Conclusion
Spatial transcriptomics offers immense potential for advancing our understanding of biological processes. However, navigating the challenges associated with input data, computational analysis, and downstream visualization is essential for extracting meaningful insights.
If you are interested in spatial omics, data ingestion, algorithm development, or visualization development, please contact info@zifornd.com.