ACMG V4 Draft Preview: Transformative Changes In Variant Classification

Image reference: / source
The field of clinical genomics continues to evolve, and with it, the guidelines for variant classification must adapt to ensure accuracy, consistency, and clarity. The Zifo Clinical Genomics team recently attended ClinGen’s workshop, Overview of DRAFT ACMG/AMP v4 Sequence Variant Guidelines, as well as the ACMG 2025 Clinical Genomics Meeting. These sessions provided valuable insights into the upcoming ACMG v4 guidelines. In this blog, we highlight key observations and updates compared to the previous version.
Key Insights
One of the most notable advancements is the complete overhaul of evidence codes assigned to each attribute. These codes have been redesigned to more intuitively reflect the core concept of each attribute, making the classification framework more concept-driven, user-friendly and precise in guiding variant classification.
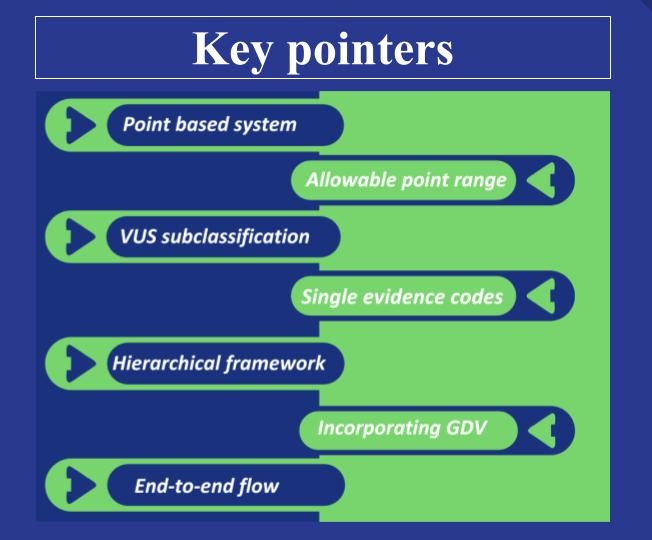
While the foundational concepts remain largely intact, the current revision has streamlined the evidence structure by consolidating overlapping codes. For example, the previously separate codes PS2 and PM6, both relating to de novo probands, have now been merged into a single unified code—OBS_DNV. This change not only simplifies the application of the guidelines but also reduces the risk of double counting evidence, thereby enhancing the accuracy of variant classification. Furthermore, unlike earlier versions where evidence codes carried fixed strengths, the new framework assigns flexible point ranges based on context, allowing for a more nuanced and dynamic evaluation.
The structure of evidence codes has also undergone a substantial evolution. The v3 evidence codes were grouped into eight separate evidence concepts, often resulting in related codes being scattered across categories without restrictions on combined usage. To address this, v4 introduces a “Hierarchical Structure of Evidence,” providing a clear framework that prevents redundancy and double counting. This hierarchy is organized into four levels: Evidence Category > Evidence Concept > Evidence Code > Code Components, each with its own sub-branching as required. A point cap system is applied across all these levels (currently under development,) to ensure that overlapping evidence does not inflate scoring. Rather than assigning fixed strengths to codes, the framework now uses continuous Bayesian point-based scoring, determined through detailed code-specific tables and flowcharts.
While the definition at the “code component” level is still under development, the high-level updates are likely to spark curiosity and surprise upon first encounter. With respect to allele frequency, v4 introduces a significant advancement by recommending customized cutoffs tailored to each gene, recognizing that gene characteristics and prevalence can vary widely. Furthermore, v4 recommends checking splicing effects for all amino acid changes and integrating functional data with predictive data. It also advances the framework by incorporating seven extensive flow diagrams, each systematically outlining end-to-end guidance detailing each step for evaluating predictive data.
Another major refinement is the heightened focus on gene-disease association as a primary criterion for reporting pathogenic variants. To classify a variant as Likely Pathogenic (LP), a minimum of moderate gene-disease association is now required. Conversely, variants associated with disputed or refuted relationships are excluded from reporting, regardless of their classification. This clear distinction improves the reliability of variant classification, ensuring that patients receive more accurate and definitive diagnoses.
One of the pivotal and most exciting advancements is the introduction of Variant of Uncertain Significance (VUS) subclassification, aimed at enhancing clarity in clinical reporting. Although detailed recommendations on this are still pending, these subcategories are designed to indicate whether a variant leans more toward benignity or pathogenicity. This added granularity supports more informed clinical decision-making and makes reporting more comprehensive. The Bayesian scale ranges from ≤ -4 to ≥10, with scores between 0 and 5 representing Uncertain Significance, further divided into Low, Mid, and High categories.
The final insight concerns the rules for combining criteria. Under v3, classification as LP required at least two attributes at an appropriate strength, regardless of the strength of individual attributes. This approach ensured a reliable assessment of pathogenicity and effectively prevented over-weighing. In contrast, the new Bayesian point-based scale, while enabling more effective combinations of evidence codes, also allows a variant to reach the LP classification with just one evidence code. This applies to benign lines of evidence as well.
Conclusion
In summary, the latest ACMG classification guidelines represent a significant advancement toward a more structured, precise, and flexible framework for variant classification. These updates are expected to greatly enhance the clarity and reliability of genetic testing, ultimately improving patient care and clinical decision-making.
However, this refined system also introduces certain challenges. It is likely to reduce the number of variants classified as VUS and increase the number of Likely Benign classifications—a shift that can be viewed as both beneficial and potentially limiting. It also places heavy reliance on predictive and functional data, with comparatively less emphasis on human observational data. Furthermore, the presence of multiple detailed flow diagrams may feel challenging for some users.
That said, the clearly defined standards offer the advantage of bringing uniformity across classifiers, reducing subjectivity, bias and variability in classification. The guidelines are currently in the pilot phase and are subject to further refinements before their official release, anticipated by mid-2026. Recognizing the critical factors discussed, the working group has sensibly recommended a year-long transition period for variant classifiers to adapt to these new changes.